|
|
DeepMILO
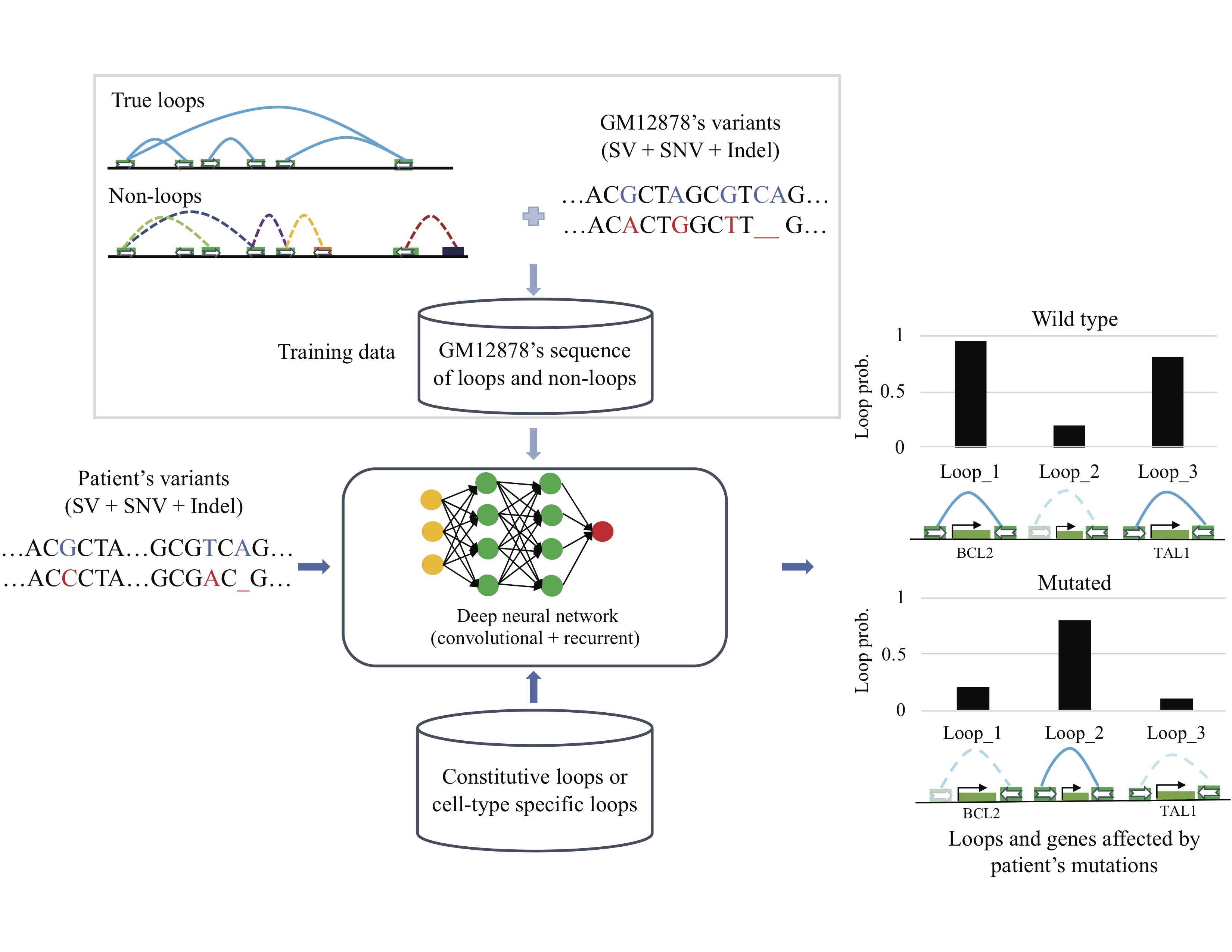
Predicting the impact of non-coding sequence variants on 3D chromatin structure using deep neural network. DeepMILO is available at DeepMILO. You can find more about this tool in Trieu et al. 2019. CNCDriver
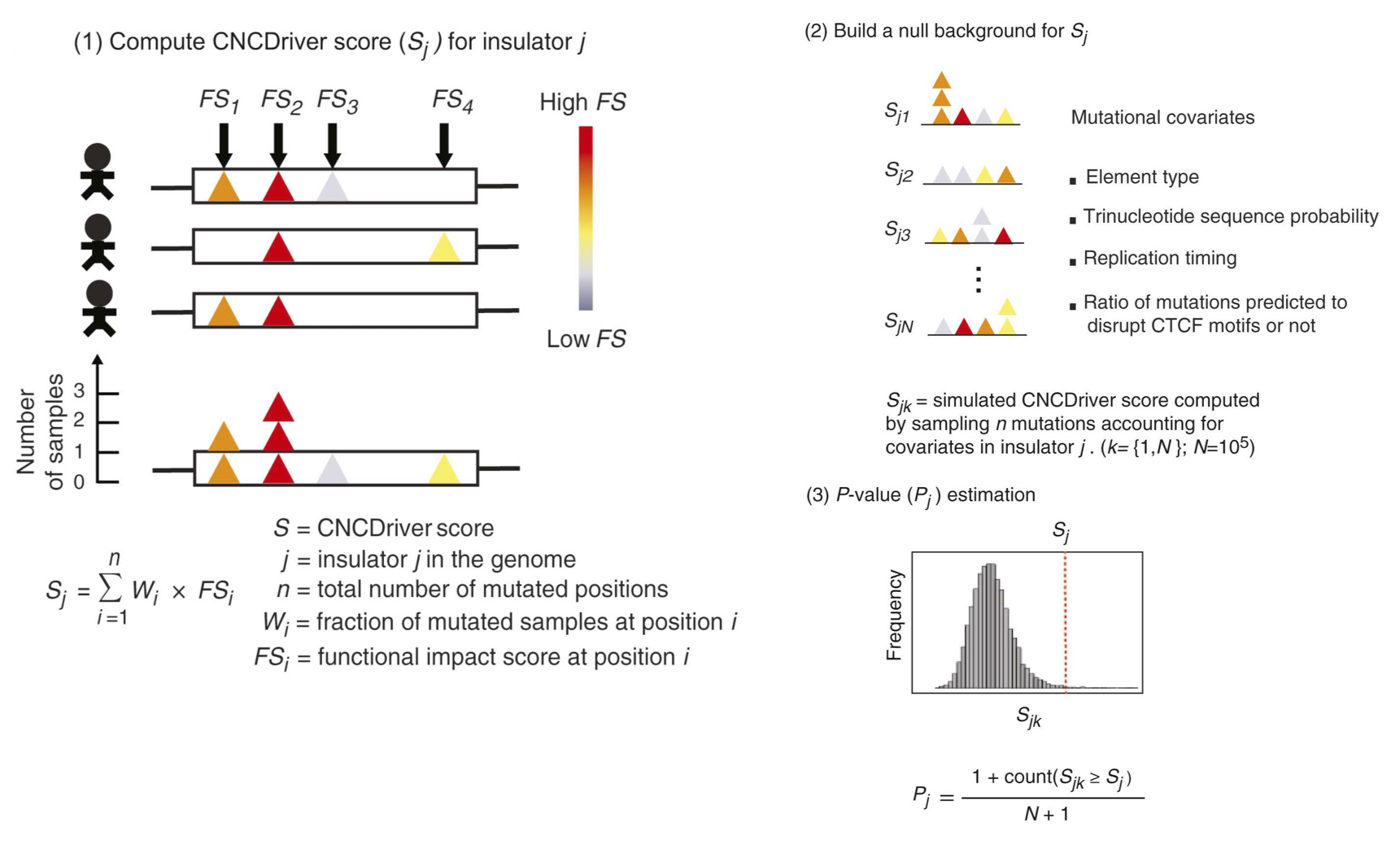
CNCDriver is a method designed to consider mutational recurrence, functional impact and mutational co-variates to identify coding and non-coding cancer driver mutations from whole-genome sequencing data. The source code of CNCDriver is available at CNCDriver, and the details of this method can be found at Liu et al. 2019. RegNetDriver
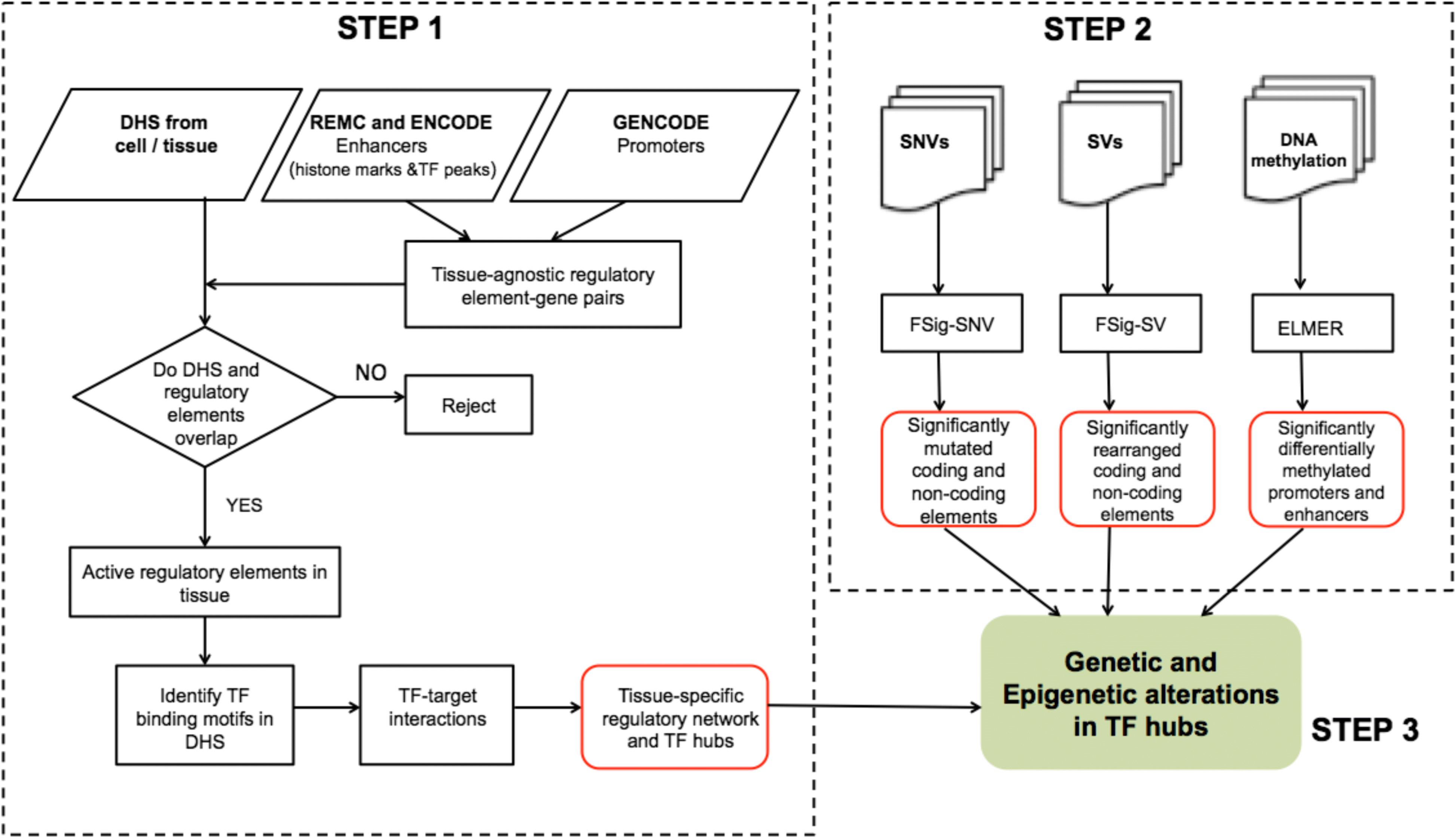
Computational method to identify tumorigenic drivers using the combined effects of coding and non-coding single nucleotide variants, structural variants, and DNA methylation changes in a DNase I hypersensitivity based regulatory network. RegNetDriver is available at RegNetDriver, and more information can be found at Dhingra et al. 2017. FunSeq2
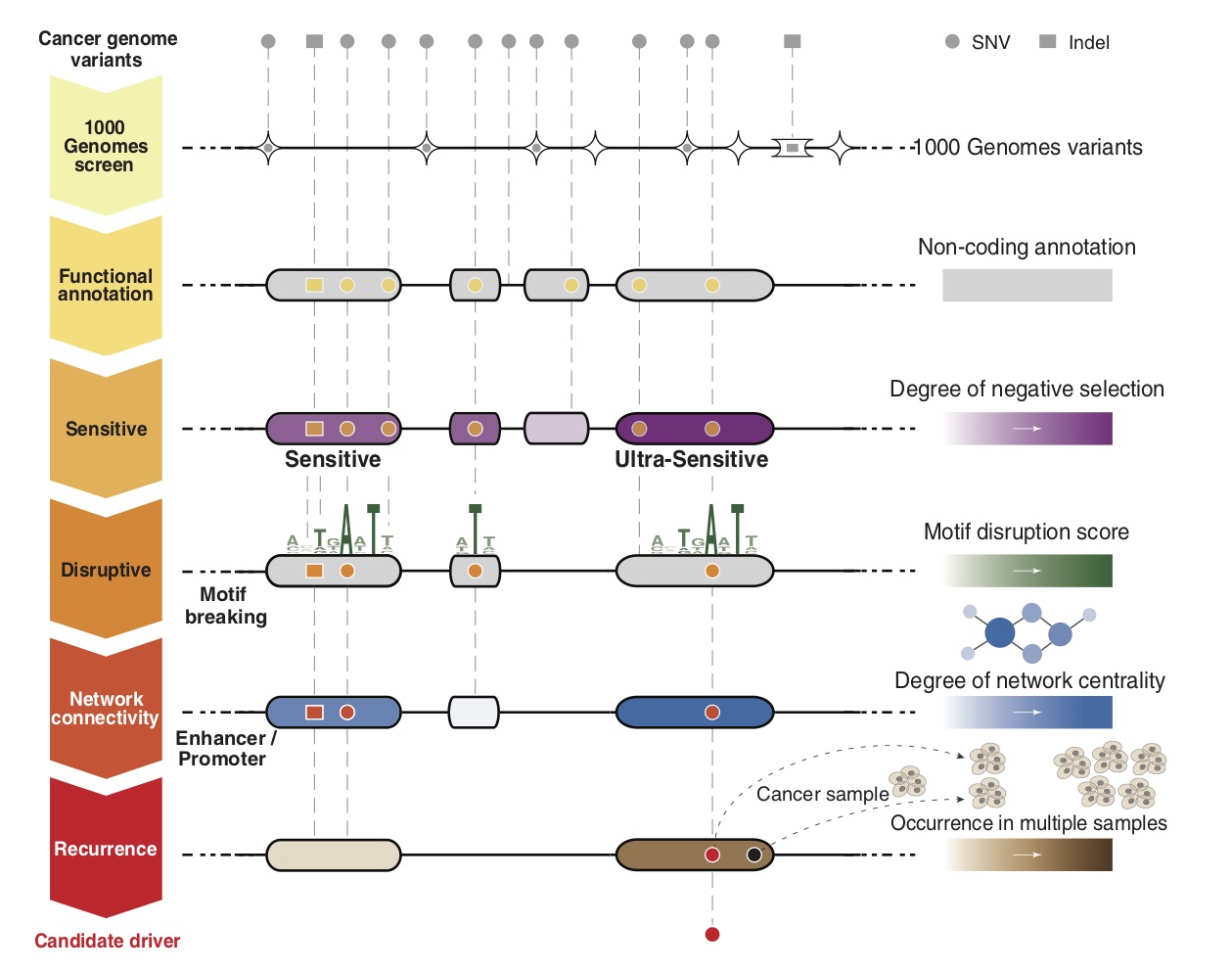
FunSeq2 is a computational framework to annotate and prioritize non-coding drivers from thousands of somatic alterations from cancer whole genomes. FunSeq2 is available at FunSeq2. You can find more about this tool in Fu et al. 2014 and Khurana et al. 2013. |